Structural basis for functional differences between Ssa and Ssb chaperone
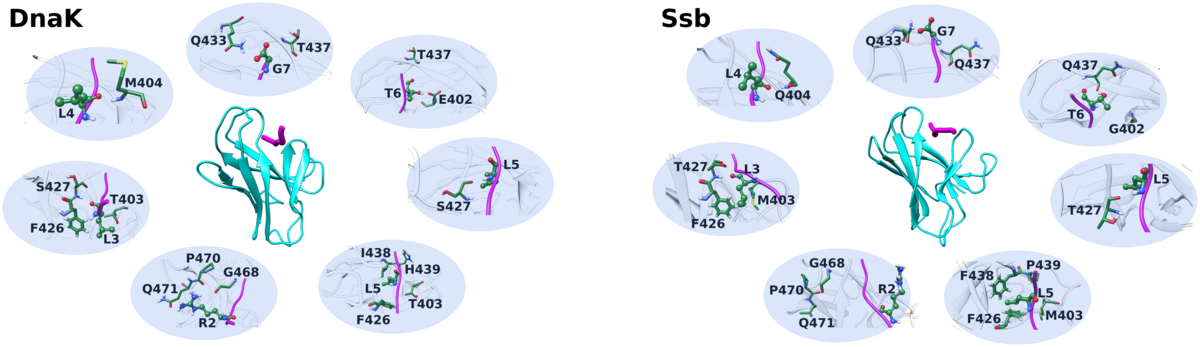
Chaperones are ubiquitous proteins that help other proteins undergo proper folding. Unlike other organisms, yeast have two distinct types of chaperones- the cytosolic Ssa and the ribosome-associated Ssb. Despite sequence and structural similarities, they function differently and the molecular basis of substrate specificity are poorly understood.
We have carried out multiple simulations of Ssa and Ssb chaperones. Only minor differences between Ssa and Ssb were observed in terms of substrate binding. However, in simulations of substrate binding domain with Lid domain attached, structural and potentially functional differences were observed between Ssa and Ssb chaperones. Our studies have identified critical residue pairs and interaction cascades that could be vital for overall structural integrity of the Lid, which seems to play an important role in ribosome-binding in Ssb. Simulation of in- silico mutants further validated our findings
Collaborator: Prof. Dr. Elke Deuerling university of Konstanz, Germany
