Structural Biology – Conjugated Proteins
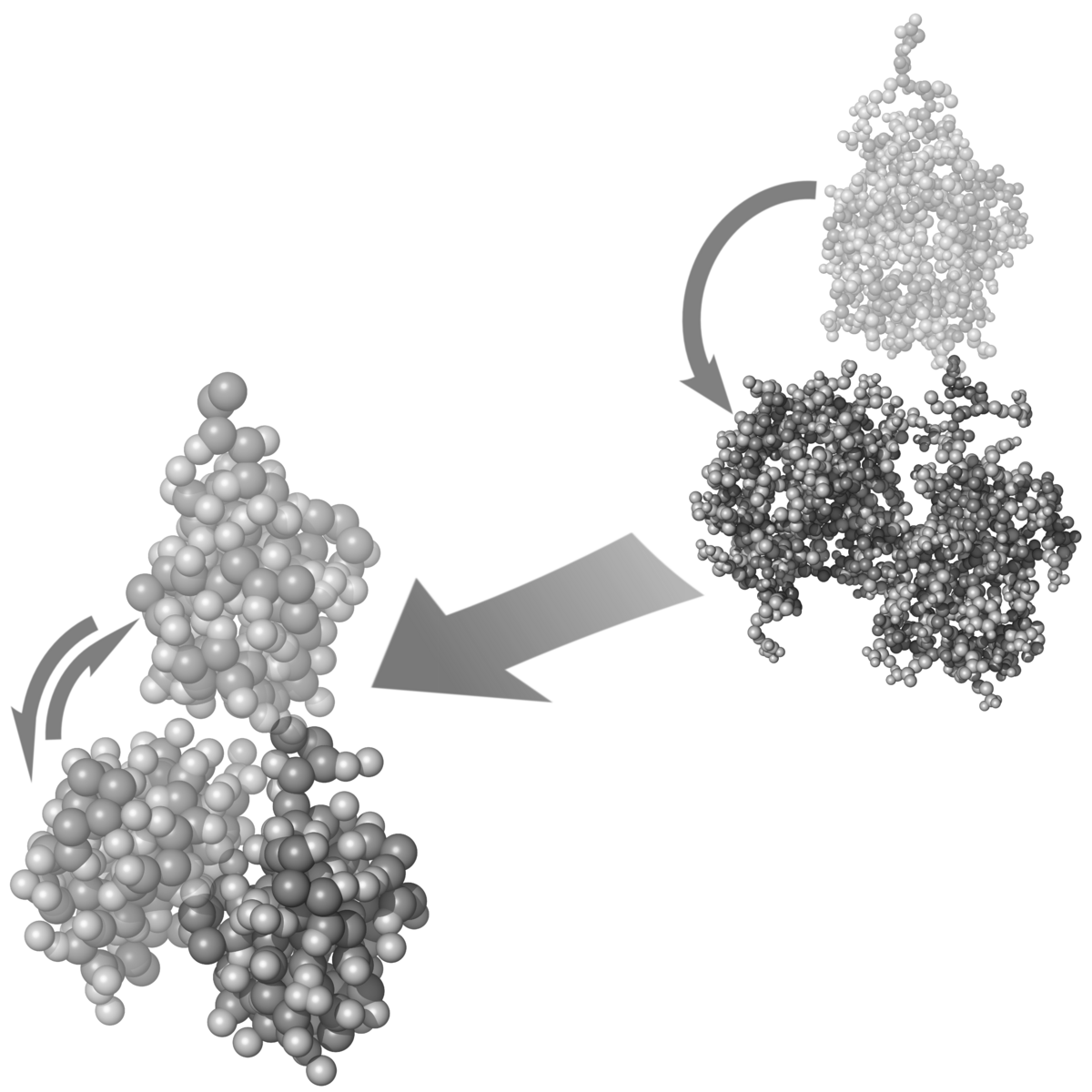
Recognition of proteins by their antibodies is enabled by their ability to fold into a well defined native structure. Molecular modelling techniques are outstanding tools to study the stability and dynamics of such structures and gain knowledge about mechanistic details on the atomistic scale. This faciliates understanding and interpretation of biological observations.
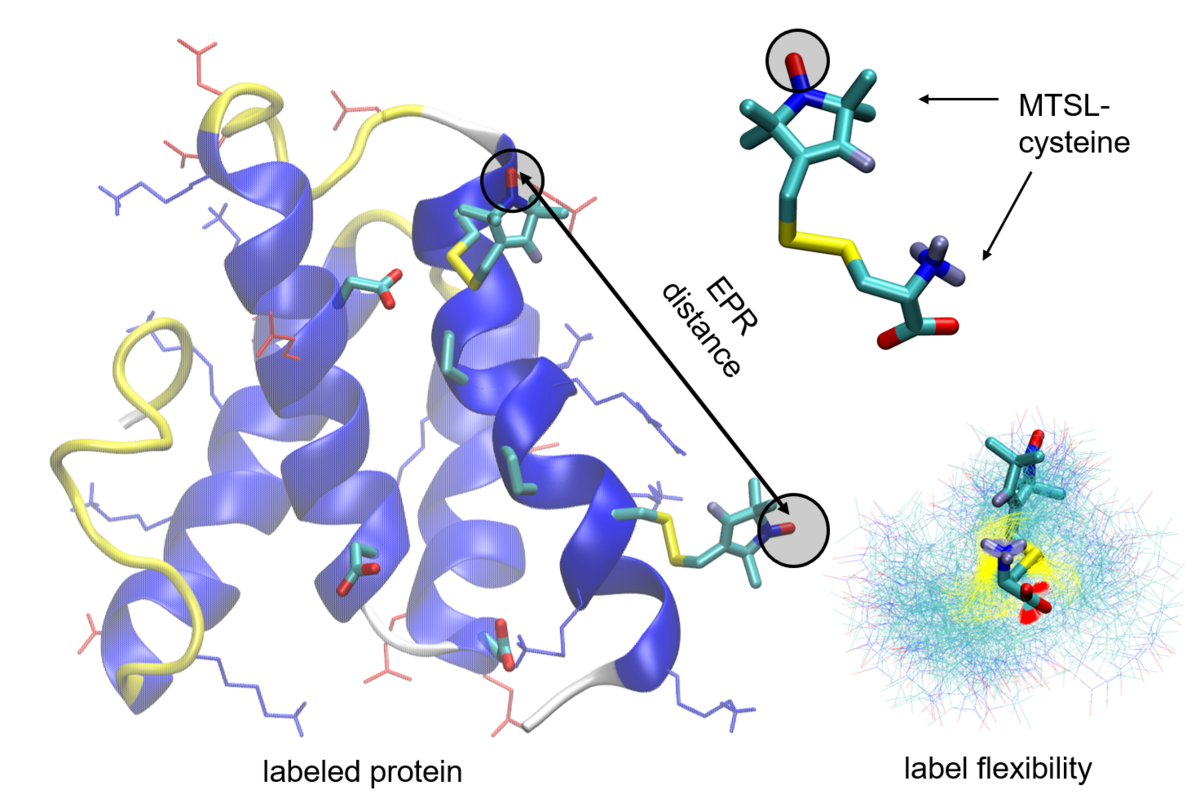
EPR-distance measurements are used to understand the role of local folding and the interaction within the ribosome-associated complex. Here especially the ribosome associated chaperone Zuotin with the Ribosome. Site directed mutagenesis and labeling with EPR-labels of the C-terminal helix bundle are utilized to probe the local folding/stability and interaction with the ribosome. Atomistic models of the protein mutants together with molecule dynamic simulations are used to rationalize the experimental studies by probing the effect of site mutations and attached EPR-label on the stability of the protein. This allows to rationalize the choice of the mutant and label positions and better understand the mechanism behind the experiment.
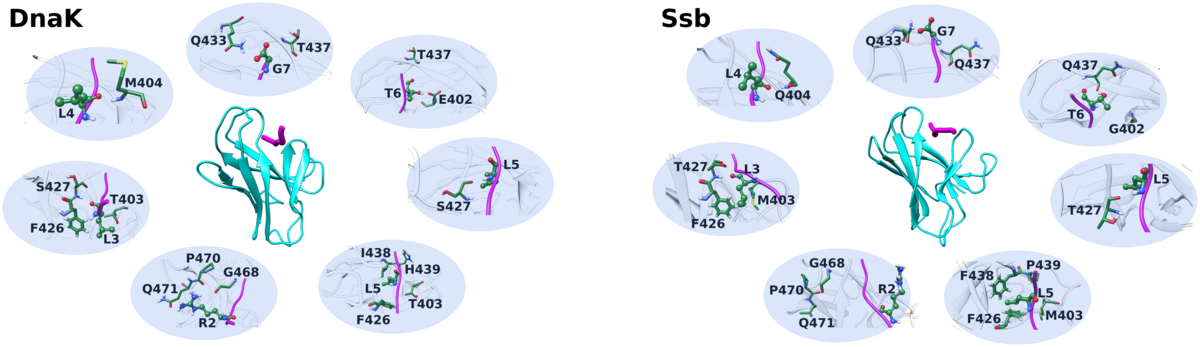
Chaperones are ubiquitous proteins that help other proteins undergo proper folding. Unlike other organisms, yeast have two distinct types of chaperones- the cytosolic Ssa and the ribosome-associated Ssb. Despite sequence and structural similarities, they function differently and the molecular basis of substrate specificity are poorly understood.
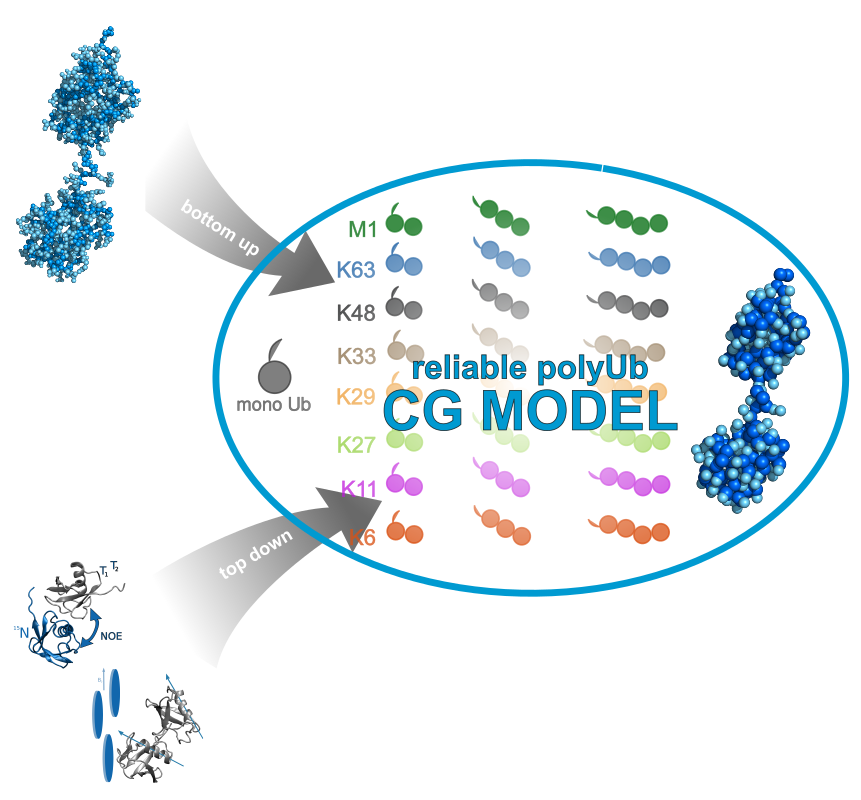
The ubiquitin system is involved in a variety of eukariotic cellular processes. It includes ubiquitilation as a post translational modification where substrate proteins are decorated with a single ubiquitin but also with chains containing several of these proteins. Different capabilities of linkage positions between distinct ubiquitin units provide variable chains. Hereinafter, ubiquitilated proteins are recognised by chain type specific antibodies and get processed by regulatory mechanisms like degradation but also by signalling and trafficking. Underlying recognition mechanisms will be elucidated by a combination of molecular modelling experiments facilitating interpretation of experimental investigations on the atomic scale.
