Model development
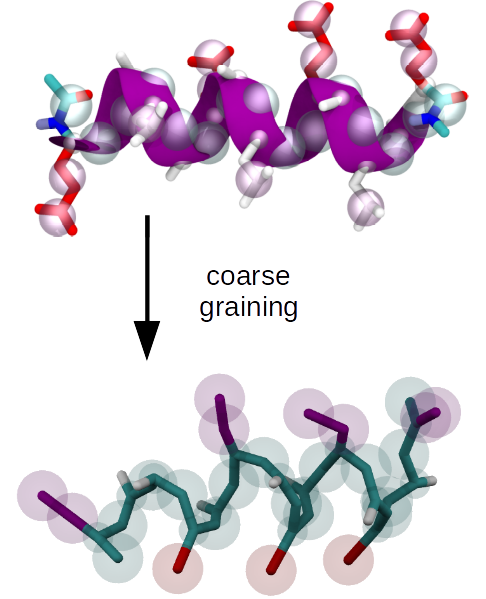
Upon developing a coarse grained (CG) model, representability and transferability limitations are a problem that's inherent to the process of reducing the number of degrees of freedom. In this context, representability refers to the question which structural or thermodynamic properties of a higher resolution reference are reproduced by the CG model, and transferability refers to the question to which extent a CG model is applicable at a state-point that differs from the one where it was parametrized.
This is naturally a highly relevant problem in simulations that involve phase transitions or structure formation processes driven by phase separation.
We investigate for several model systems how one can achieve and rationalize state-point transferability for CG models that have been parameterized in a bottom-up procedure from atomistic reference simulations, for example by choosing an appropriate reference state point.
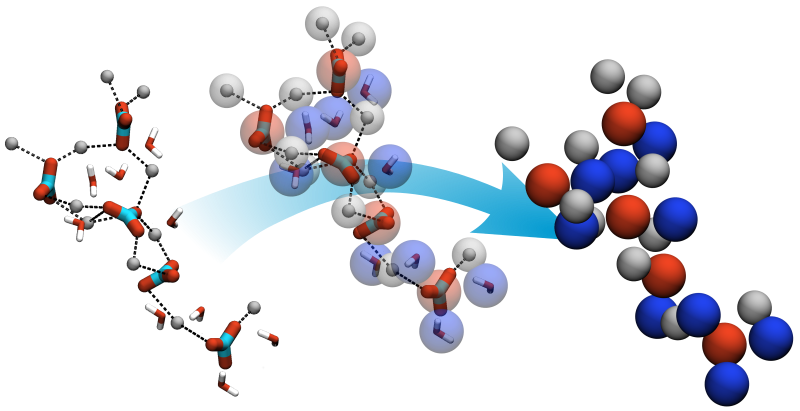
Calcium minerals can be found in various materials, like teeth, bone and shells, but also be used or occur in industrial processes. The formation of these materials can be addressed by MD simulation on different resolution levels but is computational nevertheless expensive. For insights on the early stages of nanoparticle formation and growth we develop particle-based coarse-grained models for Calcium minerals.
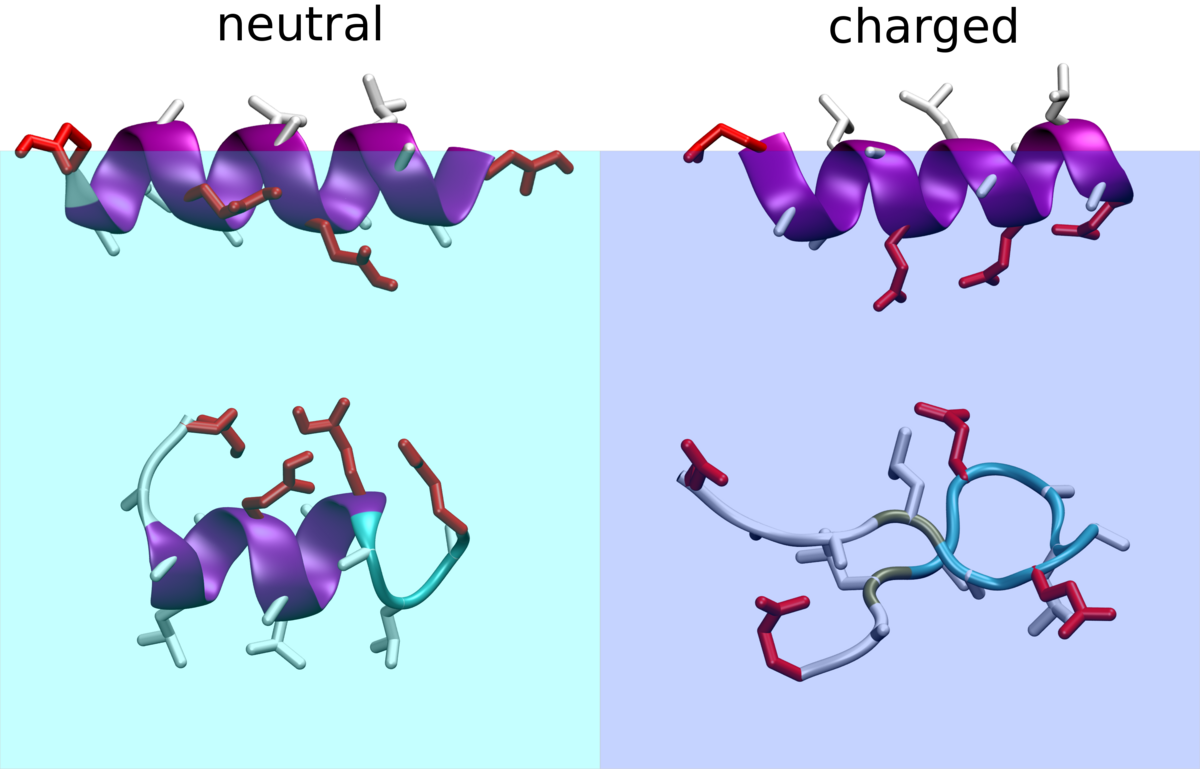
We carry out atomistic and coarse grained studies of environmentally driven folding. Amphiphilic peptides respond to the presence of interfaces, pH-changes or other changes in the solution composition by conformational transitions. Such peptides are for example initially designed as cell penetrating agents for drug targeting. Atomistic simulation studies can accompany experiments from collaborators to understand the driving forces for different folds and folding transitions. Moreover we investigate how coarse grained models can be designed such that they reproduce such responses to environment changes.
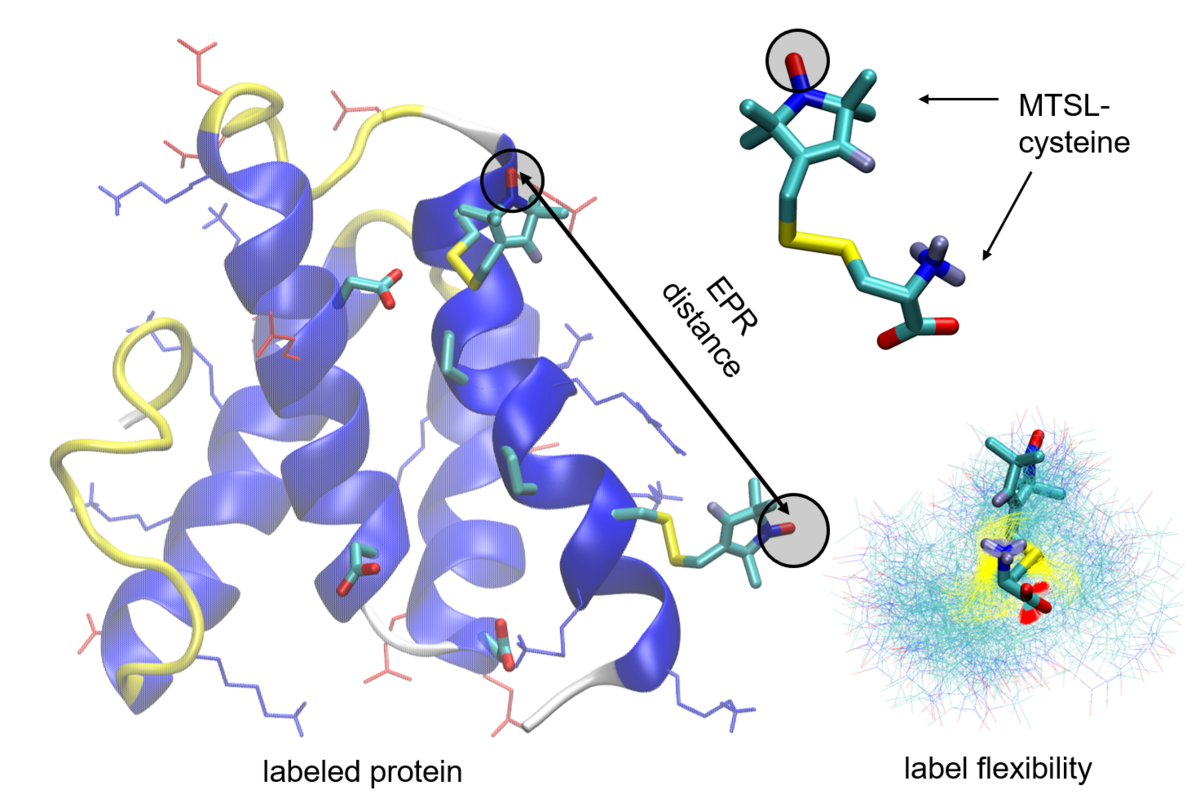
EPR-distance measurements are used to understand the role of local folding and the interaction within the ribosome-associated complex. Here especially the ribosome associated chaperone Zuotin with the Ribosome. Site directed mutagenesis and labeling with EPR-labels of the C-terminal helix bundle are utilized to probe the local folding/stability and interaction with the ribosome. Atomistic models of the protein mutants together with molecule dynamic simulations are used to rationalize the experimental studies by probing the effect of site mutations and attached EPR-label on the stability of the protein. This allows to rationalize the choice of the mutant and label positions and better understand the mechanism behind the experiment.
In recent years, machine learning techniques have become very popular and surround us in our daily life already. They help us to rank internet search results, enable software to read hand writing, recognize voice commands, and sort out spam emails. Because of the huge data amounts produced in Simulations, computational chemistry is ideal for the application of machine learning. In this project machine learning techniques are used to develop coarse grained molecule representations.
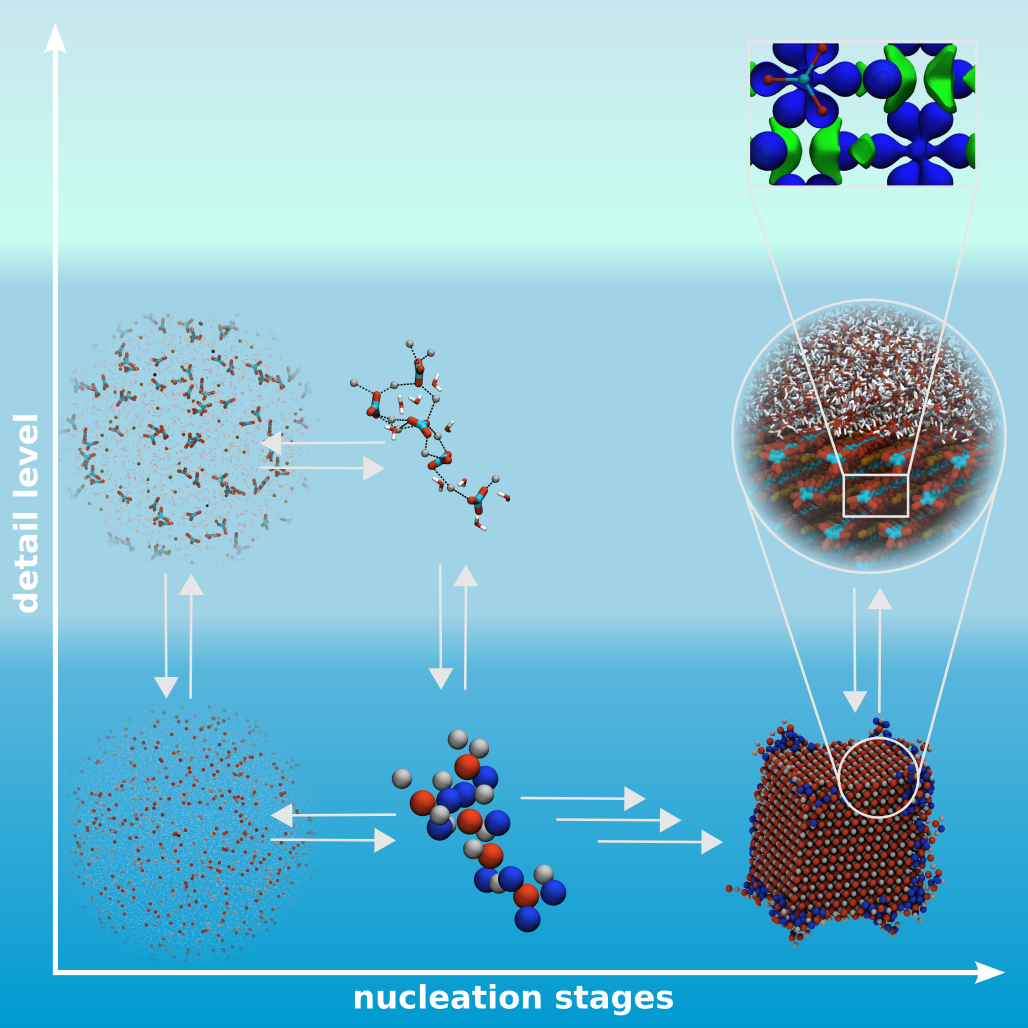
Nucleation as well as crystallization and growth of nuclei are important steps in numerous processes and applications, found in the inanimate and animated world. It could be shown that these processes can follow different pathways, classical and non-classical nucleation, which can be addressed by MD simulations. However, the different stages of these processes are on complete different time and size scales. Therefore, studies on different resolution levels, coarse-grained, atomistic and quantum mechanic levels, are needed and have to be linked to give a insight view in the underlying mechanisms.
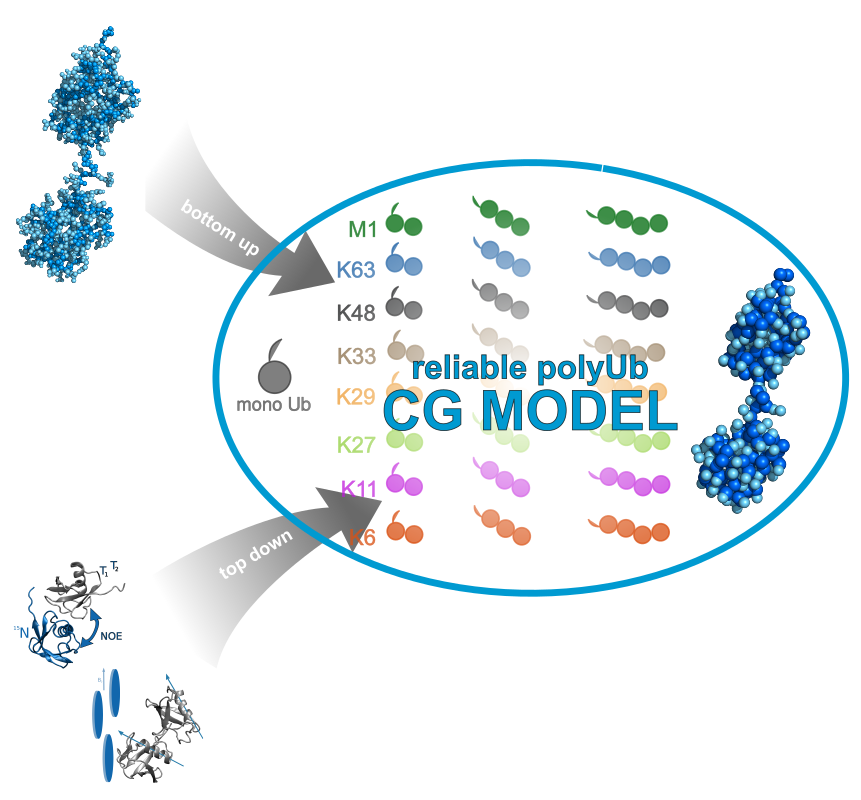
The ubiquitin system is involved in a variety of eukariotic cellular processes. It includes ubiquitilation as a post translational modification where substrate proteins are decorated with a single ubiquitin but also with chains containing several of these proteins. Different capabilities of linkage positions between distinct ubiquitin units provide variable chains. Hereinafter, ubiquitilated proteins are recognised by chain type specific antibodies and get processed by regulatory mechanisms like degradation but also by signalling and trafficking. Underlying recognition mechanisms will be elucidated by a combination of molecular modelling experiments facilitating interpretation of experimental investigations on the atomic scale.
