Riboswitches
Riboswitches reside in the 5′-UTRs of bacterial mRNAs and regulate gene expression upon interaction with small molecular ligands. Ligand binding to the aptamer domain triggers changes in the so-called expression platform, that then modulates translation initiation or transcription termination. Riboswitches are often characterized by relatively short sequences that do not require the presence of regulatory proteins, thus offering a promising general strategy for gene expression control in industrial, scientific, and future medical applications. Although the general principles are relatively well understood, mechanistic details of how ligand binding to the riboswitch exerts control on gene expression is often hard to address due to the inherent flexibility and complex dynamics of these structured RNAs.
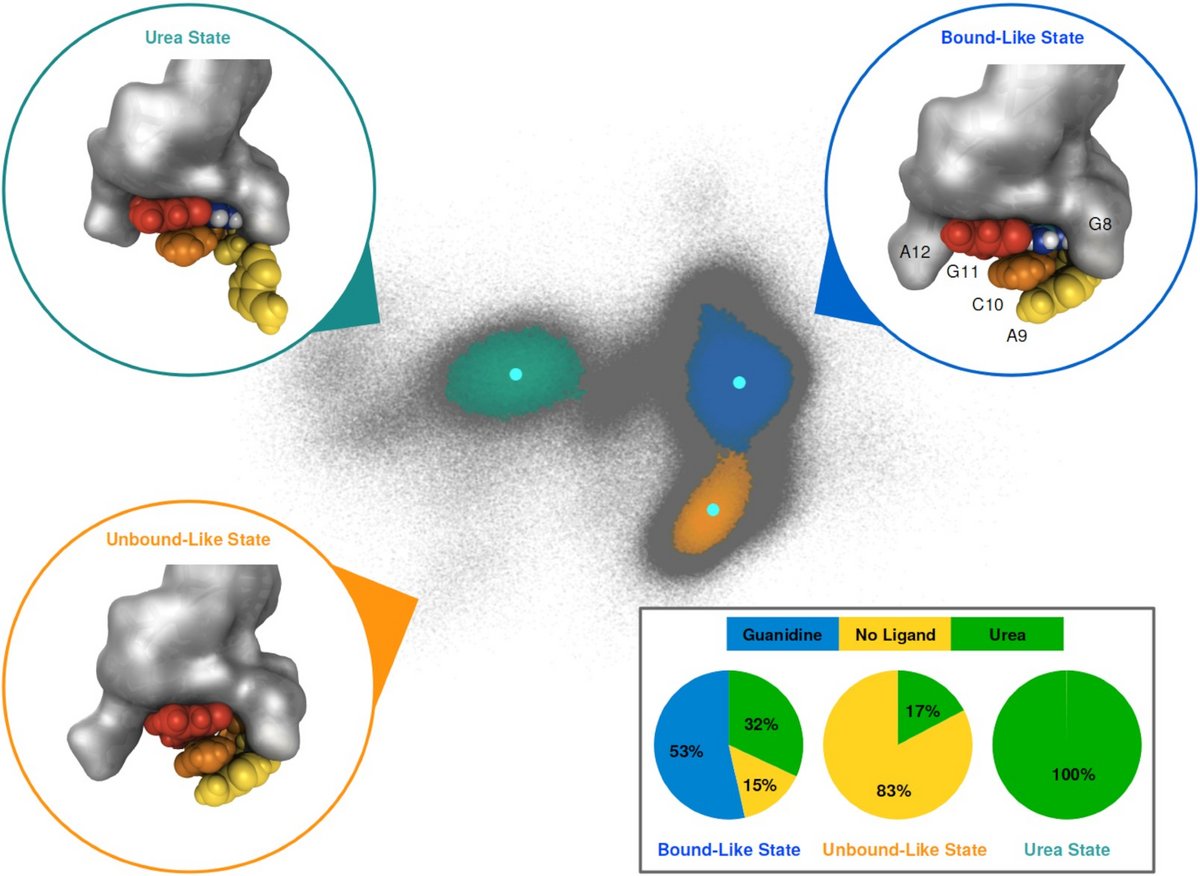
We investigate ligand binding of a member of the guanidine sensing riboswitch family, the guanidine-II riboswitch (Gd-II). It consists of two stem–loops forming a dimer upon ligand binding. Using extensive molecular dynamics simulations we have identified conformational states corresponding to ligand-bound and unbound states in a monomeric stem–loop of Gd-II and studied the selectivity of this binding.
Guanidine-II aptamer conformations and ligand binding modes through the lens of molecular simulation, Jakob Steuer, Oleksandra Kukharenko, Kai Riedmiller, Jörg S Hartig, Christine Peter, Nucleic Acids Res., gkab592, 2021. [doi]
