Model & method development
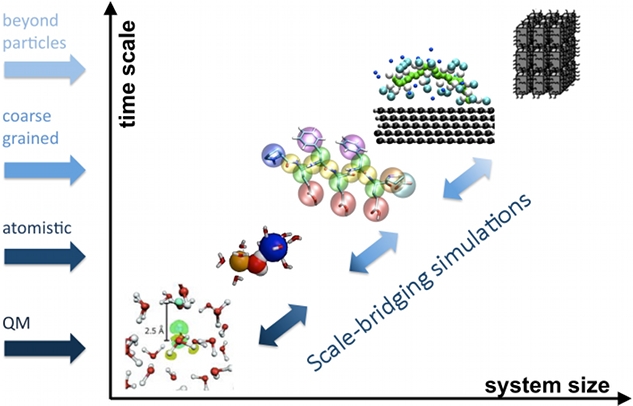
We develop systematic coarse graining methods, i.e. methods to derive models at a lower level of resolution based on information from simulations at higher, e.g. atomistic resolution.
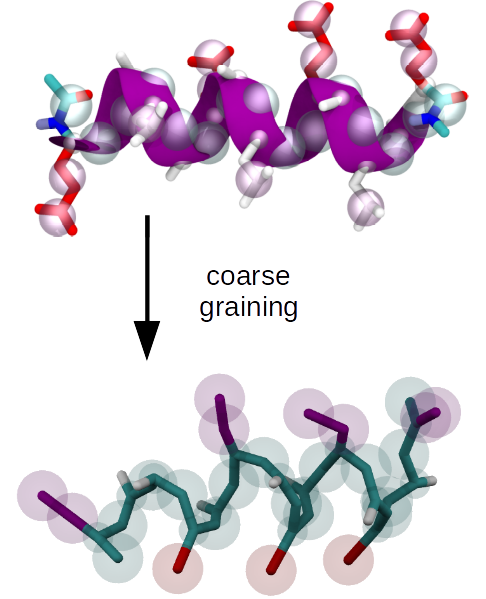
Upon developing a coarse grained (CG) model, representability and transferability limitations are a problem that's inherent to the process of reducing the number of degrees of freedom. In this context, representability refers to the question which structural or thermodynamic properties of a higher resolution reference are reproduced by the CG model, and transferability refers to the question to which extent a CG model is applicable at a state-point that differs from the one where it was parametrized.
This is naturally a highly relevant problem in simulations that involve phase transitions or structure formation processes driven by phase separation.
We investigate for several model systems how one can achieve and rationalize state-point transferability for CG models that have been parameterized in a bottom-up procedure from atomistic reference simulations, for example by choosing an appropriate reference state point.
To overcome time/length scale gaps between molecular dynamics simulations and experimental data, we improve and implement enhanced sampling techniques (HREMD, sketch-map based expansion algorithm).
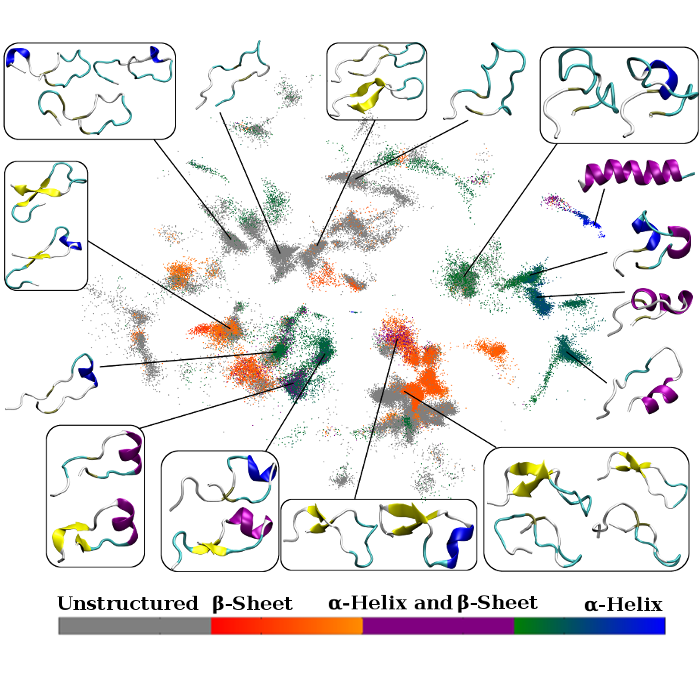
We focus on important computational issues that arise when assessing the results of MD simulations of various peptides and proteins and, more specifically, at the time of modeling their dynamical behavior, and when identifying their states and features and obtaining kinetic and thermo-dynamical estimates. We are applying and developing methods coming from machine learning. We use them for characterization of phase spaces of complex biological systems (dimensionality reduction techniques, clustering algorithms, etc.), as well as obtaining full kinetic and thermodynamical information (Markov State Models) in applications to intrinsically disordered proteins (i.e. α-synuclein, polyQ) and protein modifications (mono-ubiquitilated histone).
