Intrinsically disorderd systems
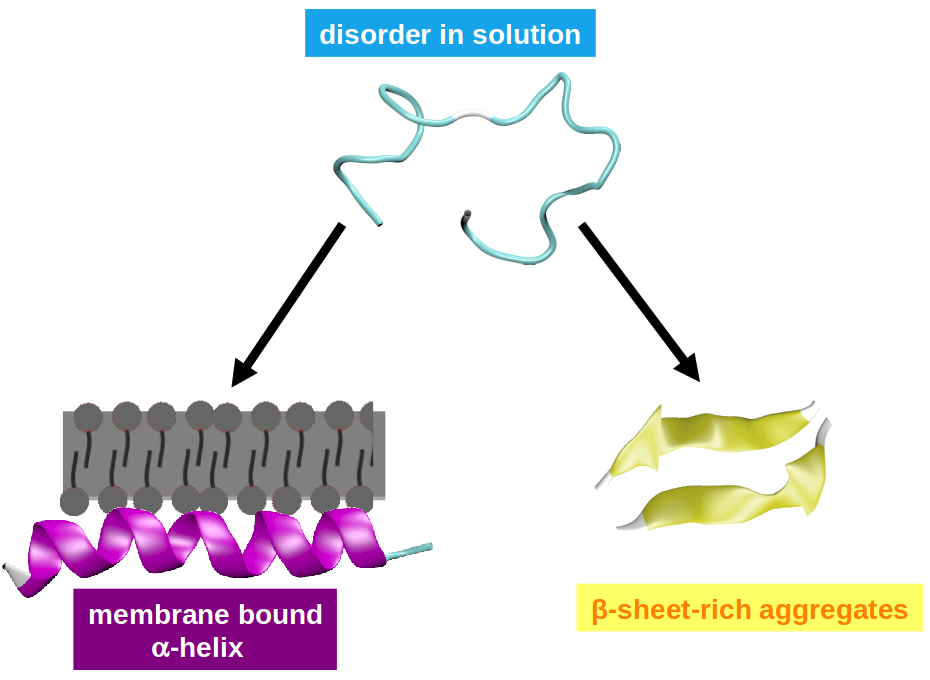
Intrinsically disordered proteins and peptides (IDPs) are biomolecules that lack well-defined secondary and tertiary structure. The great interest in this class of systems is motivated by an increasing awareness of their biological relevance. For example, several proteins that are considered intrinsically unstructured in solution such as α-synuclein, polyQ, τ-protein, and Aβ are involved in neurodegenerative diseases, such as Parkinson's disease and Alzheimer's syndrome. This is presumably due to their propensity for self-aggregation, fibril formation, but possibly also due to their interaction with biomembranes. Often, these pathophysiologically relevant phenomena are accompanied by conformational transitions where the proteins change from their disordered state into a structurally more defined β-sheet-rich or α-helical state.
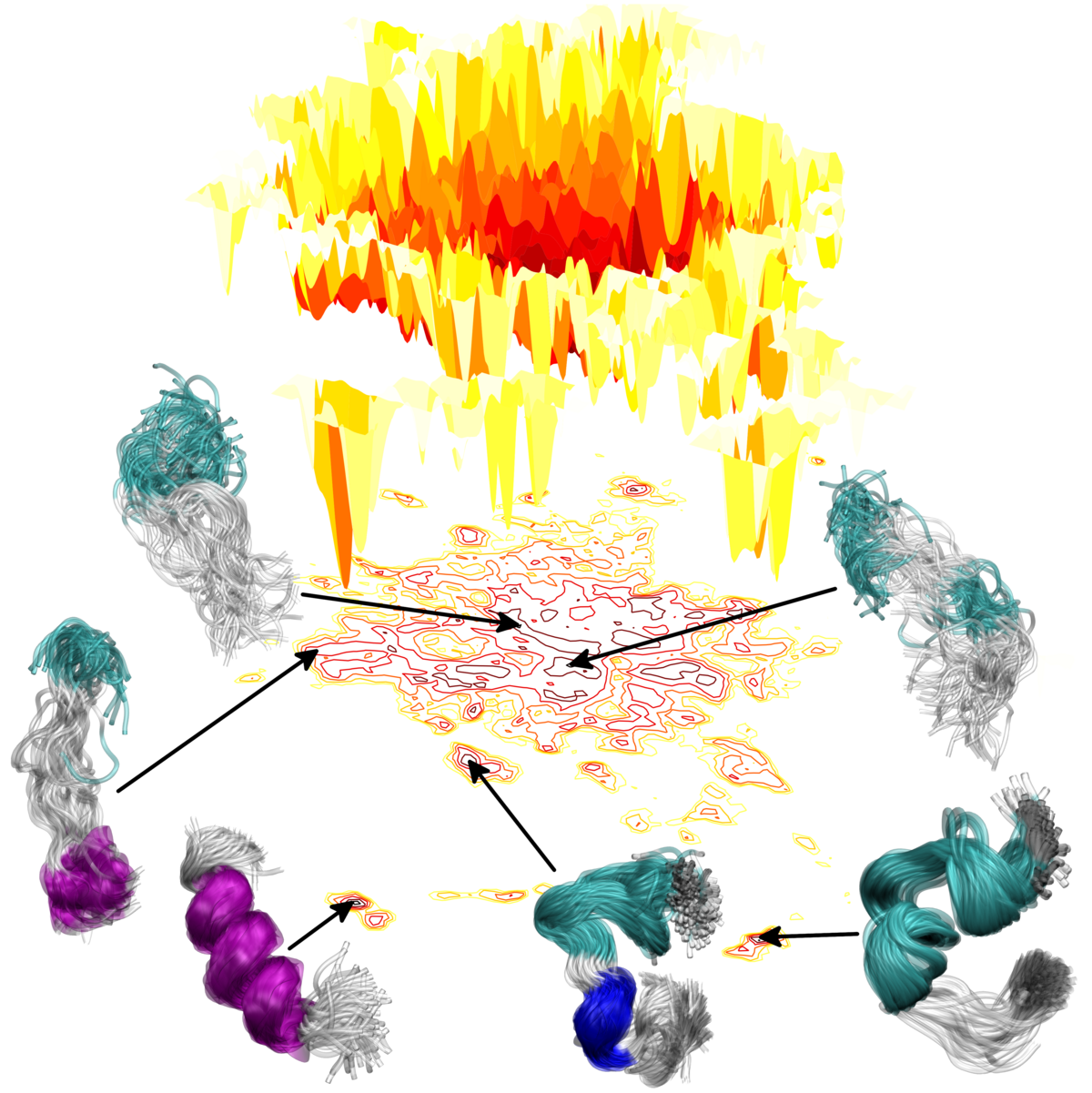
IDPs pose a challenge for many experimental methods that are traditionally used to study proteins (NMR , IR, CD, SAXS , FRET , etc.) and probe well-defined structures or secondary structures. The characteristic time scales of IDP conformational transitions, the investigation of individual molecules or the molecular mechanisms controlling protein folding upon binding remain a challenge. We are using atomistic molecular dynamics (MD) simulations to characterize the states and transitions of disordered proteins. However, it is problematic in case of IDPs. They are characterized by a comparatively shallow free energy landscape with multiple local minima lacking a dominant global minimum, resulting in a multitude of low-free energy states. In spite of comparatively low-energy barriers, their dynamics for transitions between different metastable states can be slow because of the enormity of conformational space. To obtain a convergent, statistically relevant picture of the dynamics, i.e. an accurate representation of the Boltzmann-weighted distribution of conformations, extensive sampling is required.
α-synuclein is a 140 a.a. long, presynaptic protein, abundendly found in the human brain, and although its function remains unknown, it reportedly plays a role in neurotransmitter release and synaptic function. While determining function and native structure of α-synuclein poses a challenge interesting enough in itself, it is where things go wrong - malfunction and pathological structures - which really moved α-synuclein into the spotlight of current research. Growing evidence is suggesting a causative link between the misfolding and aggregation of α-synuclein and Parkinson's Desease (PD) and other Lewy body related, neurodegenerative disorders. The accumulation of α-synuclein into amyloid fibrillar aggregates as the primary structural component in so-called Lewy bodies is a hallmark of PD.
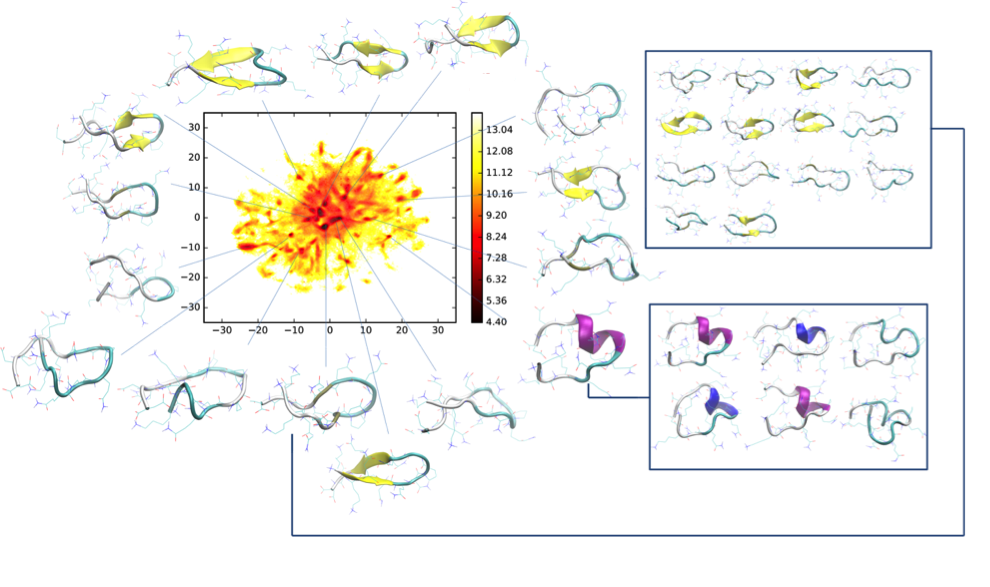
An extension of a polyglutamine (polyQ)-sequence behind a critical threshold level in some of the host proteins causes neurodegenerative diseases, the most well-known is the Huntington disease. Short polyQ-peptides are a suitable experimental and simulation model system for the study of the main pathological feature of these disaeses - aggreagtion and the processes leading to it. In addition, polyQ peptides being polar homopolymers and having large similarity between the backbone and sidechain parts of the molecule, exhibiting numerous hydrogen bonding possibilities, are an important and unique representative of the intinsically disordered systems. In this project, polyQ-peptides with various lengths and intervening residues are investigated with MD simulations coupled with advanced sampling methods and analytical techniques, in order to obtain an atomic level description of the short timescale processes happening in these conformational ensembles, the first steps of aggregation, and the effect of various factors on the complex disordered energy landscapes and aggregation, also serving as a complement to experimental data addressing larger systems and longer timescales.
