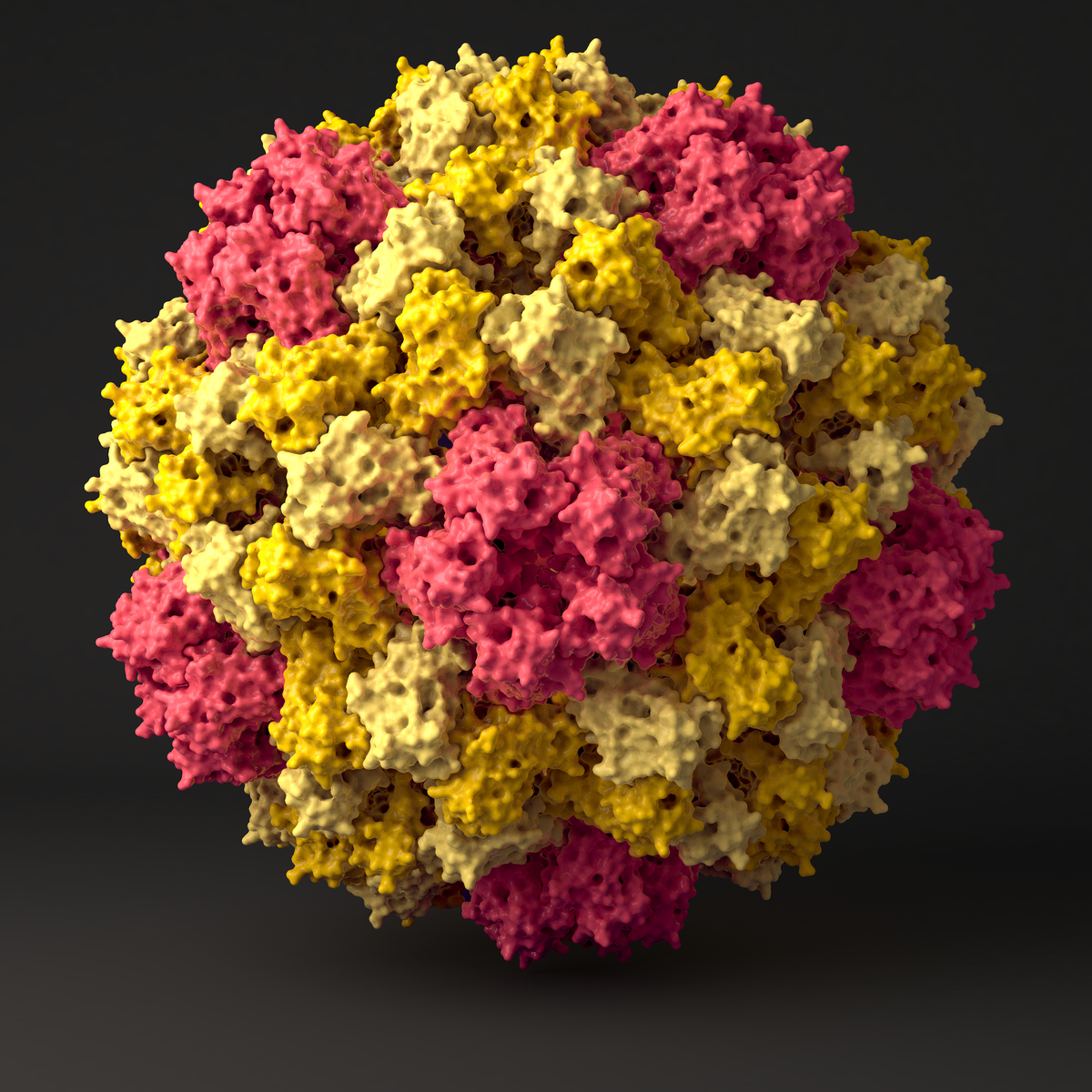
We use a combined approach of classical atomistic and different coarse grained (CG) simulation levels to investigate the 180-protein icosahedral capsid of Cowpea Chlorotic Mottle Virus (CCMV). First, the unstructured regions of the CCMV capsid proteins are studied by applying a suitable CG model together with clustering algorithms and free energy reweighting methods. The CG simulations combined with backmapping and subsequent atomistic simulations allow us to propose a multi-conformational ensemble for the experimentally- unresolved regions of the pentameric protein interface. In a second step, we use atomistic reference simulations to refine a CG protein model in such a way that it reproduces the elastic behavior of individual proteins and protein dimers in solution. The obtained model correctly predicts structural and elastic properties of bigger aggregates and mechanical properties of an entire virus capsid when compared to Atomic Force Microscopy experiment. Detailed analysis of the simulated rupture process allow us to propose an assembly model through well-defined oligomeric intermediate states, where the assembly order is regulated by the strengths of the interfacial binding, with a subsequent post-assembly reinforcement of weak spots by cooperative folding.
Aim of this project is to get a better understanding of the physical mechanisms of protein/protein and protein/lipid interactions im lipid membranes using multiscale simulations. The project is part of the Collaborative Research Center (SFB 625) in Mainz: "From Single Molecules to Nanoscopically Structured Materials". We want to study the aggregation of membrane proteins, in particular the light harvesting complex of green plants (LHC2b) - a trimeric protein that aggregates into larger scale structures, thus forming the light harvesting apparatus in the thylakoid membrane of chloroplasts. The LHC2b complex opens up questions on a wide range of levels of resolution: (i) the interaction of the chlorophyll pigments with the protein and of the protein with the lipid membrane, (ii) formation of the trimers, and (iii) the (membrane-mediated and direct) protein/protein attraction leading to the formation of nanostructures. We aim at a hierarchichal simulation approach where we connect high resolution models acting on the atomistic level with various levels of coarse grained models.
The project is a joint project with the group of Prof. Harald Paulsen at Mainz University.
