Augmenting EPR Data
Unraveling mechanistics aspects of protein function by combined approaches of elctron resonance spectroscopy an computational chemistry (modeling, docking, MD).
Docking and MD studies are utilized to understand and visualize EPR-Data obtaned for ligands bound to Pyrrolysyl-tRNA Synthetase and develop a model for its alterating action.
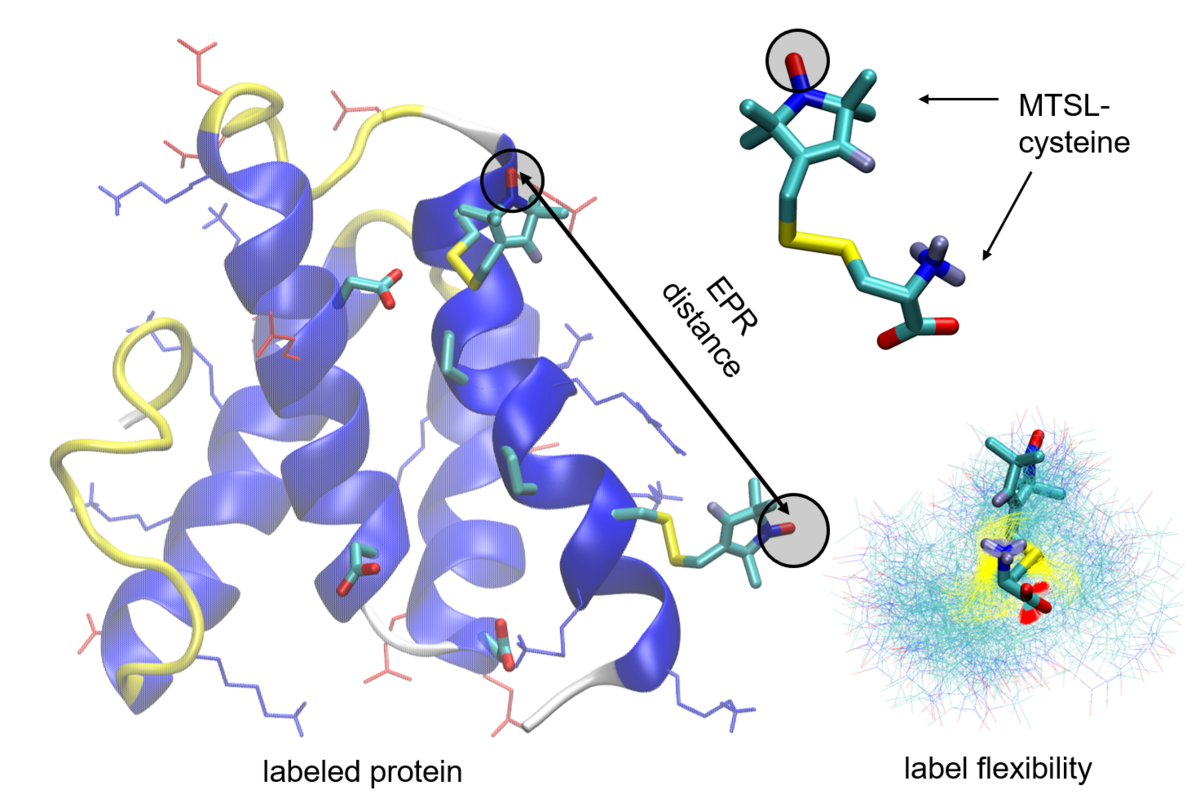
EPR-distance measurements are used to understand the role of local folding and the interaction within the ribosome-associated complex. Here especially the ribosome associated chaperone Zuotin with the Ribosome. Site directed mutagenesis and labeling with EPR-labels of the C-terminal helix bundle are utilized to probe the local folding/stability and interaction with the ribosome. Atomistic models of the protein mutants together with molecule dynamic simulations are used to rationalize the experimental studies by probing the effect of site mutations and attached EPR-label on the stability of the protein. This allows to rationalize the choice of the mutant and label positions and better understand the mechanism behind the experiment.
Deciphering molecular details of the RAC–ribosome interaction by EPR spectroscopy, S. J. Fries, T. S. Braun, C. Globisch, C. Peter, M. Drescher, E. Deuerling, Scientific Reports, 11, 8681, 2021. [doi]
