Research Interest
Genetic and Enzymatic Basis of Specialized Metabolism
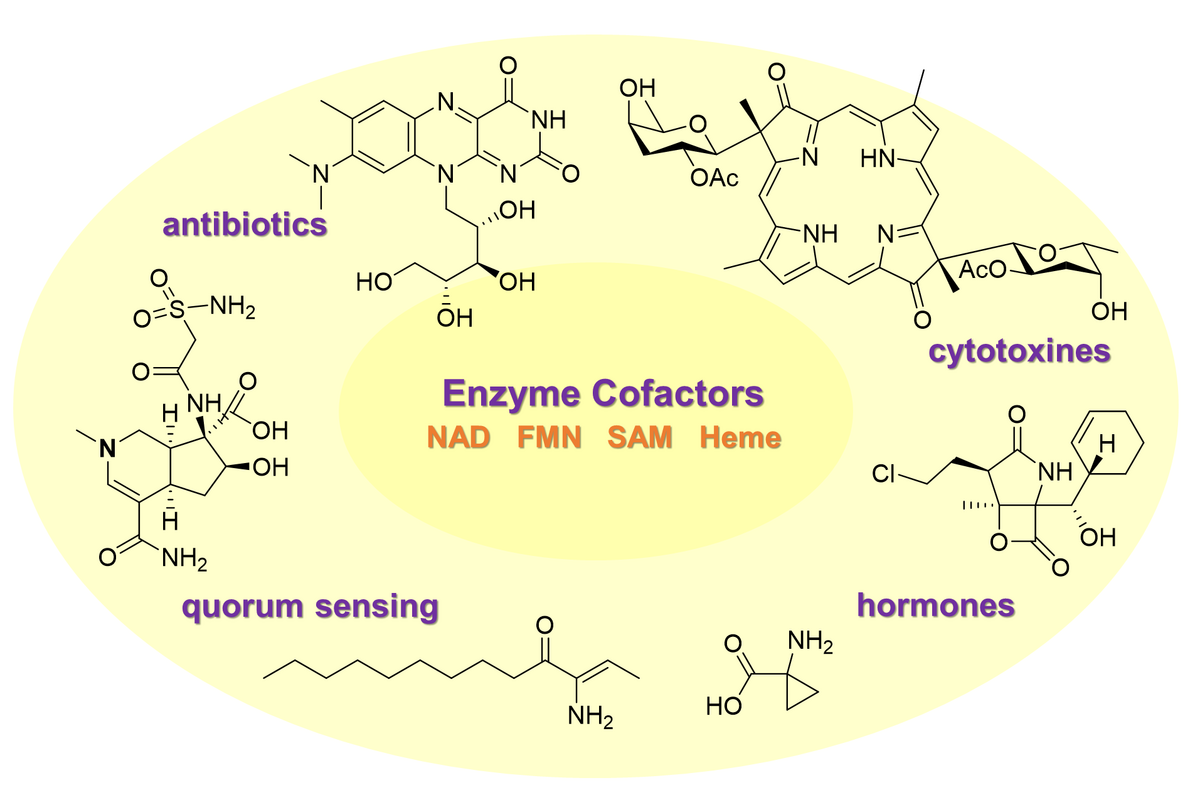
We are fascinated by Nature's ability to craft specialized metabolites with spectacular molecular architectures. These genetically encoded and evolutionary optimized small molecules exhibit potent biological activities and equip human societies to combat infectious and immunological diseases, carcinosis or help to protect crops for the food supply.
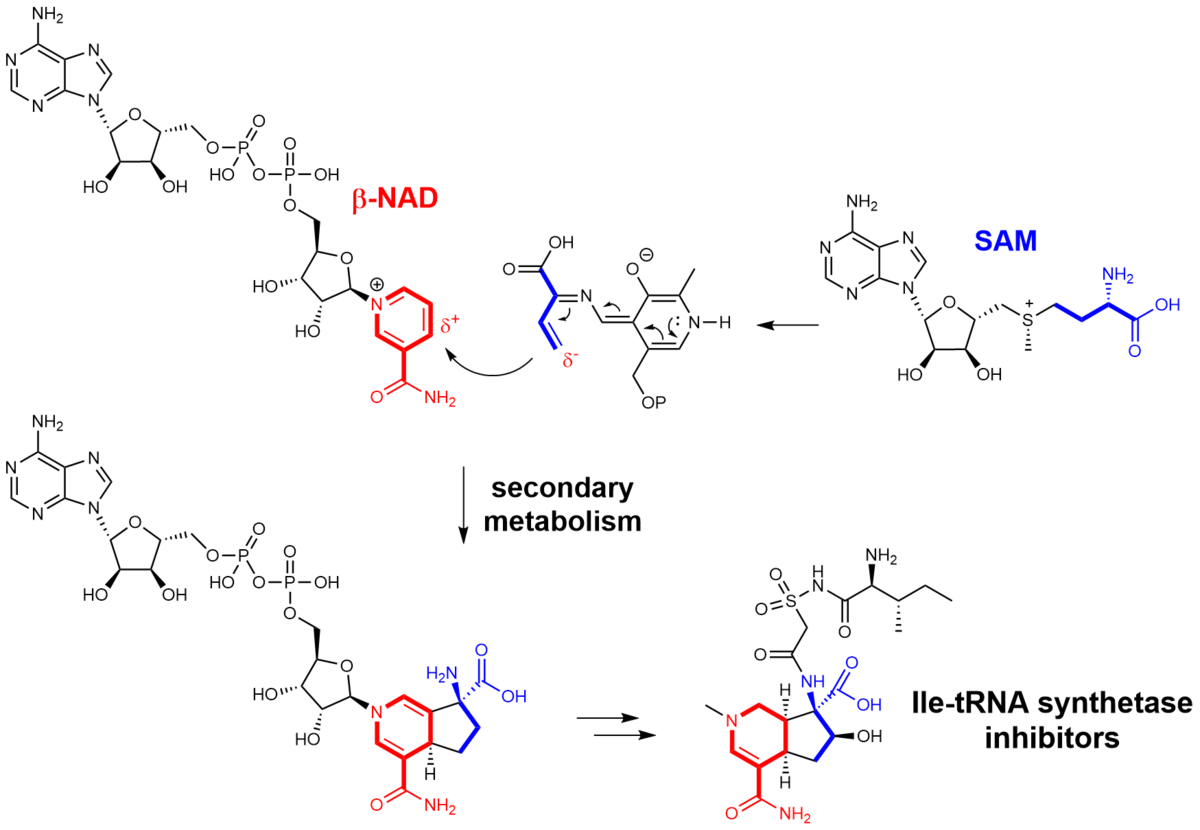
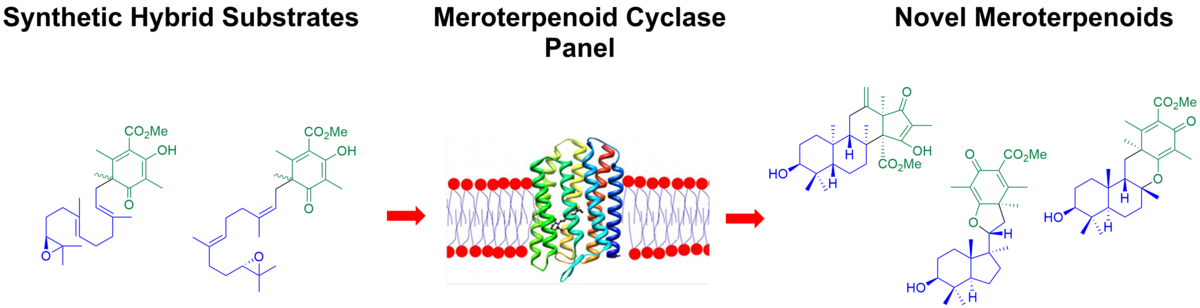
We investigate the genetic and enzymatic basis of specialized metabolism with a focus on the discovery of novel enzymes and unprecedented chemistries in non-canonical natural product pathways. Therefore, we utilize an interdisciplinary methodological approach and combine state-of-the-art techniques from organic synthesis, mass spectrometry-guided metabolomics, genomics, bioinformatics, and enzymology in order to identify novel enzymes, investigate their mechanism on a molecular level and apply the obtained knowledge for biocatalyst development and genome-based natural product discovery.
With our work, we aim to contribute to two urgent research areas with wide environmental and societal concerns: a) the development of sustainable production platforms for the synthesis of pharmaceuticals and fine chemicals, and b) the discovery of novel lead structures for antiinfectives drug development.
Funded by:

Selected Publications from postdoctoral work: